Cancer is a complex disease and developing immuno-oncology therapeutics is a challenging endeavor. Even with targeted therapies, biologics, and very specific innovative treatments, the overall probability of success for oncology drugs that enter human trials is less than four percent.1 Advances in our understanding of cancer biology, however, are helping to improve the likelihood of success. Specifically, data are accumulating on how the composition of the tumor microenvironment (TME) may affect therapeutic efficacy, underscoring the importance of profiling the TME to inform immuno-oncology drug development.
The TME is comprised of hundreds of cellular elements that influence the probability of tumor progression and the likelihood of therapeutic response or resistance. In the early stages of tumor development, the microenvironment may be immune enabling, but over time it becomes ‘polluted’ with an increasing number of adverse, immunosuppressive elements that promote tumor growth and metastasis.2
The level of immune suppression observed in human cancers is a continuum but can be categorized3 broadly as (see Figure 1):
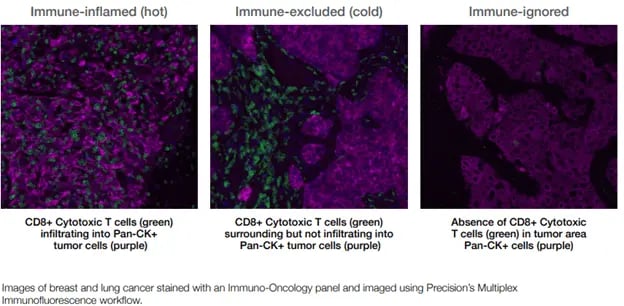 Figure 1. Immune classification of tumor microenvironments
Figure 1. Immune classification of tumor microenvironmentsImmune checkpoint inhibitors and cytotoxic immune cells only home to and infiltrate hot or very warm tumors, underscoring the importance of distinguishing among hot, cold, and immune-ignored tumors. Our current understanding is that the success of cellular therapies and treatments that rely on immune cell activation depends on:
- Proximity of the activated cells to the target tumor cells
- Effective tumor infiltration followed by activation of immune cells already within the tumor
Consequently, the spatial orientation of cells in the tumor microenvironment is medically and therapeutically important, particularly in the development of new drugs to treat solid tumors.
Using Multiplex Immunofluorescence for Spatial Profiling
Multiplex immunofluorescence (IF) allows for simultaneous detection of multiple target proteins in the same formalin-fixed paraffin-embedded (FFPE) tissue section or even the same cell, enabling higher sensitivity and a wider dynamic range than traditional chromogenic immunohistochemistry (IHC). Another advantage of this technology is the ability to generate high content data from very small and very precious or scarce clinical samples.
Multiplex IF allows for the detection of up to for identifying and phenotyping cell populations. Using software to track each cell and its associated data, it is possible to explore the spatial distribution and activation state of immune cells within a sample, which can help to distinguish hot and cold tumors and stratify patients for immunotherapy.
Precision for Medicine’s multiplex IF workflow is built upon Akoya Biosciences’ Vectra® Polaris™ Automated Quantitative Pathology System. This system integrates high content, nine-color (8-plex) multispectral imaging with the ability to easily perform six-plex (seven-color) whole-slide scanning to support the quantification and analysis of tissue sections stained with Akoya’s Opal™ detection kits using the Leica BOND RX. The resulting images are then analyzed using Akoya’s InForm or Indica Lab’s HALO® software.
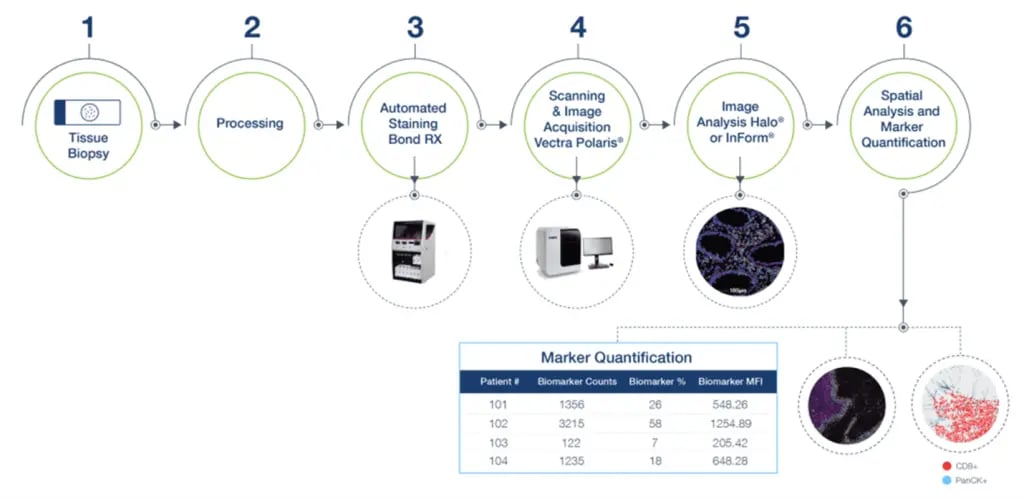 Figure 2. Precision for Medicine’s multiplex IF workflow
Figure 2. Precision for Medicine’s multiplex IF workflowIn addition to developing and validating staining panels, Precision for Medicine utilizes its expertise in IHC assay development to modify and create custom panels across various therapeutic indications. Keys to success in developing a multiplex IF panel are identifying and validating the best antibody for each target of interest and, optimizing the panel for positioning and fluorophore intensity for each target using appropriate control tissues.
Leveraging Spatial-Omics
Spatial-omics assists with the interrogation of complex, heterogenous tissues. This approach to tissue profiling reveals biological information while preserving the architecture of the tissue environment, offering the ability to understand cell characterizations without a tissue dissociation bias.
Below are a few examples of how spatial analysis can be applied to the assessment of complex cellular interactions:
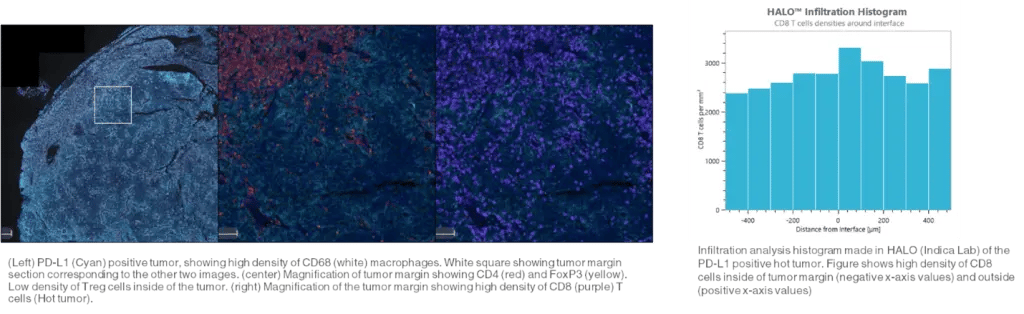 Figure 3. Infiltration analysis
Figure 3. Infiltration analysis- Infiltration analysis involves quantifying the number and depth of immune cell tumor invasion
- Single cell detection and phenotyping uses single cell intensity and spatial coordinates to study single cells or entire subpopulations and the importance of their location within the tumor microenvironment.
- Neighboring cell analysis quantifies cell-cell interactions by measuring the distance of each cell of one phenotype to its closest neighbor of a different phenotype.
Learn more about Precision’s multiplex IF services >
Key Takeaway
Knowledge of the tumor microenvironment must be incorporated into clinical trials to increase the overall success rate of investigative oncology drugs. Understanding the phenotypes and spatial arrangement of cells present in the tumor microenvironment will help developers to better guide therapy and better predict outcomes.
References
1. Wong CH, Siah KW, Lo AW. Estimation of clinical trial success rates and related parameters. Biostatistics. 2019;20(2):273-286.
2. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557-4566.




