Precision Medicine and the Rising Importance of High-Quality Biospecimens
Human biospecimens are essential for advancing scientific research across the entire continuum of preclinical and clinical drug development. In this era of precision medicine, high-quality biospecimens play a critical role as essential resources for target identification, biomarker discovery research, and therapeutic development strategies. They play a pivotal role in assay development and validation, enabling researchers to conduct molecular and cellular analysis that inform personalized treatment strategies.
Here, we focus on the use of biospecimens for tissue-based analyses at different stages of development, leveraging real-life examples to demonstrate how well-annotated samples can be used to generate actionable data from bench to bedside and back, creating endless value.
The Biospecimen Journey: From Preclinical Histology to Clinical Studies in Drug Development
In preclinical development, it is common to apply histology, immunohistochemistry (IHC), and genomic analysis to interrogate tissues or biopsy samples. These techniques allow researchers to gain comprehensive insight into disease biology, pathophysiology, and the molecular mechanisms underpinning various conditions. This information is critical for drug discovery and development processes.
Alzheimer’s Disease Case Study: The Impact of Quality Biospecimens
Precision for Medicine was engaged to develop a well-characterized tissue microarray (TMA) to support groundbreaking Alzheimer’s disease (AD) research, utilizing quality-controlled and ethically sourced biospecimens. In addition to our tissue analysis capabilities and deep experience in supporting central nervous system (CNS) disease research, Precision for Medicine has access to a range of sample types across a range of CNS diseases through a strong, longstanding partnership with a European brain bank.
| Access To Diseased CNS Tissues | CNS Tissue Analysis Capabilities |
|---|---|
| • Alzheimer’s Disease • Parkinson’s Disease • Frontotemporal dementia • Dementia – Lewy Body Variant • Multi system atrophy • Multiple Sclerosis • Amyotrophic lateral sclerosis • Progressive Supranuclear Palsy • Pick’s Disease • Pain |
• mRNA expression analysis -Quantitative RT-PCR -In situ hybridization (RNAscope) • Protein expression analysis -Immunohistochemistry -Multiplexed immunofluorescence -Western blot (automated – Biotechne JESS) • Protein isolation / enrichment -PHF-Tau -Alpha-synuclein |
The objective of the TMA was to investigate target and biomarker expression across the staged progression of AD. To achieve this, we worked collaboratively with our brain bank partner to source temporal cortex tissue from patients with early (Braak I and II) , mid (Braak III and IV), and late (Braak V and VI) stage AD and select samples based on additional factors such as amyloid score, apolipoprotein E (APOE) genotype data, cerebrospinal fluid (CSF) pH, and clinical history. We then created a TMA comprising tissues from five individual donors per stage and five non-disease donors.
-

Clinical Trials - Biomarkers - Clinical Trial Strategy
Advancing the Use of Biomarkers in CNS Drug Discovery and Development
- |
The resulting TMA was qualified using immunohistochemistry (IHC) for expression of hallmark disease markers: phosphorylated tau (phospho-tau or p-tau) and amyloid beta (Aβ) 1-42, a hyperphosphorylated form of Aβ (see Figure 1). Both p-tau and Aβ 1-42 expression were found to increase with Braak stage in the TMA.

To enhance the clinical relevance of the study, the TMA was further characterized by investigating two glial markers, IBA-1 for microglia and GFAP for astrocytes, highlighting the role of quality biospecimens in neurodegenerative disease research. Both microglia and astrocytes undergo changes in frequency and morphology in different stages of AD, as seen for both IBA-1and GFAP in Figure 2.
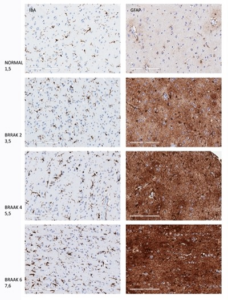 Figure 2. Changes in staining intensity and cell morphology at different stages of AD
Figure 2. Changes in staining intensity and cell morphology at different stages of AD
To take it even further, digital pathology and AI-powered image analysis allow simultaneous evaluation of staining patterns in matched fields of view. These advanced technologies enable the precise detection and quantification of potential targets and biomarkers, allowing for deeper insights into their association with specific cell populations. This is a fundamental step in translational medicine, bridging laboratory findings to clinical applications. (see Figure 3).

Figure 3. Evaluation of staining patterns in matched fields of view shows overlapping distribution of hallmark disease and glial markers and dense aggregation of GFAP immunoreactive astrocytes in the location of a suspected plaque
Case Study: Immuno-Oncology and the Detailed Interrogation of Tissue with Multiplex Immunofluorescence (mIF)
Complex diseases and target organ structures often require extremely detailed interrogation of the tissue to yield insights into disease biology or assess drug efficacy or safety. While tissue biopsies are invasive, they enable localized analysis of protein biomarkers and topographic investigation of disease- and drug-related changes. In recent years, spatial analysis of multiple tissue biomarkers using multiplex immunofluorescence (mIF) has become routine for generating or supporting exploratory data during clinical development, especially in conditions with core immune components, such as cancer and inflammatory or autoimmune diseases.
Importantly, mIF maximizes that data that can be extracted from a very limited amount of tissue, whether from a preclinical animal model or a clinical biopsy sample. mIF can interrogate multiple markers on a single slide, generative quantitative data on cell counts, intensity, co-localization, and even spatial localization.
For instance, Precision for Medicine developed multiplex immunofluorescence (mIF) panels to meticulously characterize various mouse tumor models, demonstrating the power of precise tissue analysis in oncology research. Using the assays developed, we were able to characterize post-treatment changes in target expression and in the tumor microenvironment using animal model samples and then translate those learnings to human tissue-based assays to support exploratory biomarker evaluation in early clinical development.
Transitioning to a Clinical Assay: Considerations and Challenges
Transitioning from preclinical tissue-based analyses to assays that might be more applicable in the clinical development setting requires consideration of several factors, including:
- Accessible sample types
- The sample collection process
- The logistics of transporting samples from multiple clinical sites to either a central or specialty lab for downstream analysis
In CNS conditions, a key driver for assay development is the need for either less- or non-invasive means for biomarker analysis, diagnosis, patient selection or stratification, or disease monitoring. While CSF is an optimal source for biomarkers given its direct relation to the extracellular space in the brain, it is more invasive than imaging- or blood-based approaches. In AD, modalities such as improved positron emission tomography (PET) imaging tracers for detecting Aβ and tau pathology and blood- or exosome-based biomarkers are well-suited for clinical applications. Researchers are also:
- Utilizing data garnered from AD biomarker measurements in the brain to identify predictive blood biomarkers and even correlate those blood biomarker levels with cognitive function
- Examining biomarker combinations that can differentiate between stages of AD with greater specificity and sensitivity
- Developing machine learning techniques from CSF proteomic profiles to identify potential biomarkers that can predict rates of cognitive decline
Development of any of these assays, especially those leveraging liquid biopsies, to correlate back to brain pathology relies on access to the right biospecimens. These specimens, which must be ethically sourced and rigorously quality-controlled, support basic research, proof of concept studies, and the broader goals of personalized medicine and targeted therapies.
In the immuno-oncology space, Precision for Medicine developed a fit-for-purpose assay for immune cell profiling in non-small cell lung cancer (NSCLC). To qualify this assay, we looked at tissue samples from three individual donors with NSCLC, using three slides from each donor and investigated three matched regions of interest per slide to assess consistency of both the cell counts and the percentage of target-expressing cells. We were able to demonstrate that intra-donor data were highly consistent for the targets of interest (see Figure 4) and the assay was appropriate to apply to a clinical trial cohort.
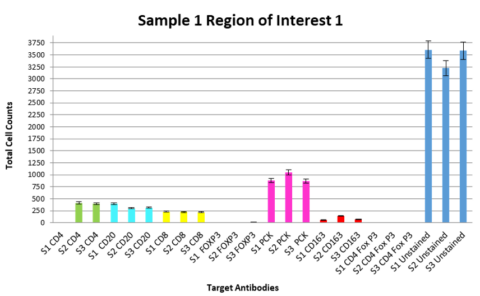
Given that there is inter-individual variability in tumor target expression, patient selection and predictive biomarkers may require large tissue cohorts for establishment of an expression level cut point. Being able to readily access well-annotated samples to support these assay validations is imperative.
Case Study: Custom IHC Assay Development for Clinical Trial Enrollment in Oncology
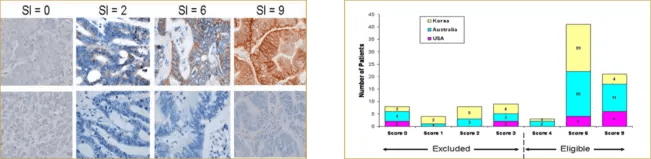
To support enrollment in a global Phase 1 oncology study, Precision for Medicine developed and validated a custom IHC assay, reagent kit, and scoring index (SI) for assessing P-cadherin, a cellular adhesion protein that contributes to oncogenesis. The SI was based on the tumor area stained multiplied by the staining intensity (see Figure 5). Patients whose tumor biopsies had an SI of 4 or higher were eligible for enrollment.
Expanding Access to Well-Characterized Biospecimens for Assay Validation
Validation of clinical trial assays requires the right biospecimens. Critical bottlenecks in the assay validation process are:
- Finding and accessing samples positive for the biomarker of interest
- Getting orthogonal confirmation for positive and negative calls
In 2020, Precision for Medicine launched the Precision Oncology Sequencing Initiative (Project POSI), a next generation sequencing (NGS) program focused on genetically characterizing as many of the millions of oncology FFPE samples and histology slides in our biorepository as possible and banking these characterized samples for future use. We have adopted a platform- and assay-agnostic approach to sequencing, instead aiming to leverage the most useful and appropriate technology for generating oncology insights.
Through partnerships, Precision for Medicine has characterized nearly 12,000 FFPE samples across over 15 cancer indications. More than 10,000 FFPE samples across 11 oncology indications have been characterized using, a targeted amplicon-based 50-gene panel that screens for single nucleotide variants (SNVs), indels, fusions, and amplifications. An additional 800 FFPE samples are being sequenced per month using a pan-cancer 500+ gene capture-based panel for tumor mutational burden (TMB), microsatellite instability (MSI), SNVs, indels, copy number variations (CNVs), fusions, and splice variants for verification, validation, and generation of orthogonal data for regulatory submissions.
Precision for Medicine also has approximately 350 donors enrolled in a liquid biopsy sequencing initiative, which involves prospective collections under IRB-approved protocols that allow us to perform repeat or longitudinal biopsies as additional needs arise.
For researchers, having access to our comprehensive and easily searchable database of ethically sourced and quality-controlled biospecimens, complete with NGS characterization and associated variant data, significantly streamlines the assay development process in precision medicine. Precision for Medicine also offers objective third-party orthogonal testing services for validating inter-laboratory reproducibility to support enrollment, predictive biomarkers, and companion diagnostic development.
The Future of Pathology: Digital Solutions and Artificial Intelligence (AI)
With the volume of pathology work required to support target and biomarker discovery and assay validation, keeping track of all the data generated and having a robust audit trail is crucial. At Precision for Medicine, we have onboarded Halo AP from Indica Labs to manage our pathology data and workflow. This cloud-based solution also enables multiple pathologists to collaborate, independent of geography.
Taking it a step further, pathology data sets like ours are highly sought after for training artificial intelligence (AI) and machine learning. To that end, we have scanned hematoxylin and eosin (H&E)-stained slides of every NGS sample to supplement the physical tissue block, residual extracted DNA and RNA, variant data, and demographic information available. These high-resolution digital pathology images serve as robust training sets for machine learning algorithms, unlocking the potential to detect novel targets and biomarkers, thereby automating the discovery process and transitioning it into cloud-based platforms for global collaboration.
Conclusion: The Integrated Approach of Precision for Medicine
Precision for Medicine is a global precision medicine clinical research organization (CRO) that integrates clinical trial execution, specialty assay development, custom central lab services, advanced data sciences, and comprehensive biospecimen management solutions. With a focus on oncology, neurology, and complex diseases, we are committed to advancing patient-centric healthcare through cutting-edge molecular diagnostics and targeted treatments. We have seven labs globally, delivering a range of biomarker assays that interrogate the full spectrum of analytes in biofluids, cells, and tissues.
We also have millions of IRB-approved, clinically-annotated biospecimens and actively manage custom prospective collections across multiple sample types and formats, uniquely positioning us to offer an end-to-end solution for integrating biomarkers and biospecimens from concept to clinic to cloud.
.webp?width=593&height=366&name=PFM-Figure-6.-The-pivotal-role-of-high-quality-biospecimens-throughout-the-continuum-of-development-593x366%20(1).webp)
Figure 6. The pivotal role of high quality biospecimens throughout the continuum of development
Discover how our high-quality biospecimens are the key to driving successful therapeutic and diagnostic development. Watch our on-demand webinar now and take the next step towards groundbreaking healthcare innovation.





